Missing values
Missing values aren’t a data type as such, but are an important concept in R; the way different functions handle missing values can be both helpful and frustrating in equal measure.
Missing values in a vector are denoted by the letters NA, but notice that these letters are unquoted. That is to say NA is not the same as "NA"!
To check for missing values in a vector (or dataframe column) we use the is.na() function:
nums.with.missing <- c(1, 2, NA)
nums.with.missing
[1] 1 2 NA
is.na(nums.with.missing)
[1] FALSE FALSE TRUEHere the is.na() function has tested whether each item in our vector called nums.with.missing is missing. It returns a new vector with the results of each test: either TRUE or FALSE.
We can also use the negation operator, the ! symbol to reverse the meaning of is.na. So we can read !is.na(nums) as “test whether the values in nums are NOT missing”:
# test if missing
is.na(nums.with.missing)
[1] FALSE FALSE TRUE
# test if NOT missing (note the exclamation mark in front of the function)
!is.na(nums.with.missing)
[1] TRUE TRUE FALSEWe can use the is.na() function as part of dplyr filters:
airquality %>%
filter(is.na(Solar.R)) %>%
head(3) %>%
pander| Ozone | Solar.R | Wind | Temp | Month | Day |
|---|---|---|---|---|---|
| NA | NA | 14.3 | 56 | 5 | 5 |
| 28 | NA | 14.9 | 66 | 5 | 6 |
| 7 | NA | 6.9 | 74 | 5 | 11 |
Or to select only cases without missing values for a particular variable:
airquality %>%
filter(!is.na(Solar.R)) %>%
head(3) %>%
pander| Ozone | Solar.R | Wind | Temp | Month | Day |
|---|---|---|---|---|---|
| 41 | 190 | 7.4 | 67 | 5 | 1 |
| 36 | 118 | 8 | 72 | 5 | 2 |
| 12 | 149 | 12.6 | 74 | 5 | 3 |
Complete cases
Sometimes we want to select only rows which have no missing values — i.e. complete cases.
The complete.cases function accepts a dataframe (or matrix) and tests whether each row is complete. It returns a vector with a TRUE/FALSE result for each row:
complete.cases(airquality) %>%
head
[1] TRUE TRUE TRUE TRUE FALSE FALSEThis can also be useful in dplyr filters. Here we show all the rows which are not complete (note the exclamation mark):
airquality %>%
filter(!complete.cases(airquality))
Ozone Solar.R Wind Temp Month Day
1 NA NA 14.3 56 5 5
2 28 NA 14.9 66 5 6
3 NA 194 8.6 69 5 10
4 7 NA 6.9 74 5 11
5 NA 66 16.6 57 5 25
6 NA 266 14.9 58 5 26
7 NA NA 8.0 57 5 27
8 NA 286 8.6 78 6 1
9 NA 287 9.7 74 6 2
10 NA 242 16.1 67 6 3
11 NA 186 9.2 84 6 4
12 NA 220 8.6 85 6 5
13 NA 264 14.3 79 6 6
14 NA 273 6.9 87 6 8
15 NA 259 10.9 93 6 11
16 NA 250 9.2 92 6 12
17 NA 332 13.8 80 6 14
18 NA 322 11.5 79 6 15
19 NA 150 6.3 77 6 21
20 NA 59 1.7 76 6 22
21 NA 91 4.6 76 6 23
22 NA 250 6.3 76 6 24
23 NA 135 8.0 75 6 25
24 NA 127 8.0 78 6 26
25 NA 47 10.3 73 6 27
26 NA 98 11.5 80 6 28
27 NA 31 14.9 77 6 29
28 NA 138 8.0 83 6 30
29 NA 101 10.9 84 7 4
30 NA 139 8.6 82 7 11
31 NA 291 14.9 91 7 14
32 NA 258 9.7 81 7 22
33 NA 295 11.5 82 7 23
34 78 NA 6.9 86 8 4
35 35 NA 7.4 85 8 5
36 66 NA 4.6 87 8 6
37 NA 222 8.6 92 8 10
38 NA 137 11.5 86 8 11
39 NA 64 11.5 79 8 15
40 NA 255 12.6 75 8 23
41 NA 153 5.7 88 8 27
42 NA 145 13.2 77 9 27Sometimes it’s convenient to use the . (period) to represent the output from the previous pipe command. For example, we could rewrite the previous example as:
airquality %>%
filter(!complete.cases(.)) # note the . (period) here in place of `airmiles`
Ozone Solar.R Wind Temp Month Day
1 NA NA 14.3 56 5 5
2 28 NA 14.9 66 5 6
3 NA 194 8.6 69 5 10
4 7 NA 6.9 74 5 11
5 NA 66 16.6 57 5 25
6 NA 266 14.9 58 5 26
7 NA NA 8.0 57 5 27
8 NA 286 8.6 78 6 1
9 NA 287 9.7 74 6 2
10 NA 242 16.1 67 6 3
11 NA 186 9.2 84 6 4
12 NA 220 8.6 85 6 5
13 NA 264 14.3 79 6 6
14 NA 273 6.9 87 6 8
15 NA 259 10.9 93 6 11
16 NA 250 9.2 92 6 12
17 NA 332 13.8 80 6 14
18 NA 322 11.5 79 6 15
19 NA 150 6.3 77 6 21
20 NA 59 1.7 76 6 22
21 NA 91 4.6 76 6 23
22 NA 250 6.3 76 6 24
23 NA 135 8.0 75 6 25
24 NA 127 8.0 78 6 26
25 NA 47 10.3 73 6 27
26 NA 98 11.5 80 6 28
27 NA 31 14.9 77 6 29
28 NA 138 8.0 83 6 30
29 NA 101 10.9 84 7 4
30 NA 139 8.6 82 7 11
31 NA 291 14.9 91 7 14
32 NA 258 9.7 81 7 22
33 NA 295 11.5 82 7 23
34 78 NA 6.9 86 8 4
35 35 NA 7.4 85 8 5
36 66 NA 4.6 87 8 6
37 NA 222 8.6 92 8 10
38 NA 137 11.5 86 8 11
39 NA 64 11.5 79 8 15
40 NA 255 12.6 75 8 23
41 NA 153 5.7 88 8 27
42 NA 145 13.2 77 9 27This is nice because we can apply the complete.cases function to the output of the previous pipe. For example, if we wanted to select complete cases for a subset of the variables we could write:
airquality %>%
select(Ozone, Solar.R) %>%
filter(!complete.cases(.))
Ozone Solar.R
1 NA NA
2 28 NA
3 NA 194
4 7 NA
5 NA 66
6 NA 266
7 NA NA
8 NA 286
9 NA 287
10 NA 242
11 NA 186
12 NA 220
13 NA 264
14 NA 273
15 NA 259
16 NA 250
17 NA 332
18 NA 322
19 NA 150
20 NA 59
21 NA 91
22 NA 250
23 NA 135
24 NA 127
25 NA 47
26 NA 98
27 NA 31
28 NA 138
29 NA 101
30 NA 139
31 NA 291
32 NA 258
33 NA 295
34 78 NA
35 35 NA
36 66 NA
37 NA 222
38 NA 137
39 NA 64
40 NA 255
41 NA 153
42 NA 145Or alternatively:
rows.to.keep <- !complete.cases(select(airquality, Ozone, Solar.R))
airquality %>%
filter(rows.to.keep) %>%
head(3) %>%
pander| Ozone | Solar.R | Wind | Temp | Month | Day |
|---|---|---|---|---|---|
| NA | NA | 14.3 | 56 | 5 | 5 |
| 28 | NA | 14.9 | 66 | 5 | 6 |
| NA | 194 | 8.6 | 69 | 5 | 10 |
Missing data and R functions
It’s normally good practice to pre-process your data and select the rows you want to analyse before passing dataframes to R functions.
The reason for this is that different functions behave differently with missing data.
For example:
mean(airquality$Solar.R)
[1] NAHere the default for mean() is to return NA if any of the values are missing. We can explicitly tell R to ignore missing values by setting na.rm=TRUE
mean(airquality$Solar.R, na.rm=TRUE)
[1] 185.9315In contrast some other functions, for example the lm() which runs a linear regression will ignore missing values by default. If we run summary on the call to lm then we can see the line near the bottom of the output which reads: “(7 observations deleted due to missingness)”
lm(Solar.R ~ Temp, data=airquality) %>%
summary
Call:
lm(formula = Solar.R ~ Temp, data = airquality)
Residuals:
Min 1Q Median 3Q Max
-169.697 -59.315 6.224 67.685 186.083
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -24.431 61.508 -0.397 0.691809
Temp 2.693 0.782 3.444 0.000752 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 86.86 on 144 degrees of freedom
(7 observations deleted due to missingness)
Multiple R-squared: 0.07609, Adjusted R-squared: 0.06967
F-statistic: 11.86 on 1 and 144 DF, p-value: 0.0007518Normally R will do the ‘sensible thing’ when there are missing values, but it’s always worth checking whether you do have any missing data, and addressing this explicitly in your code
Patterns of missingness
The mice package has some nice functions to describe patterns of missingness in the data. These can be useful both at the exploratory stage, when you are checking and validating your data, but can also be used to create tables of missingness for publication:
mice::md.pattern(airquality) 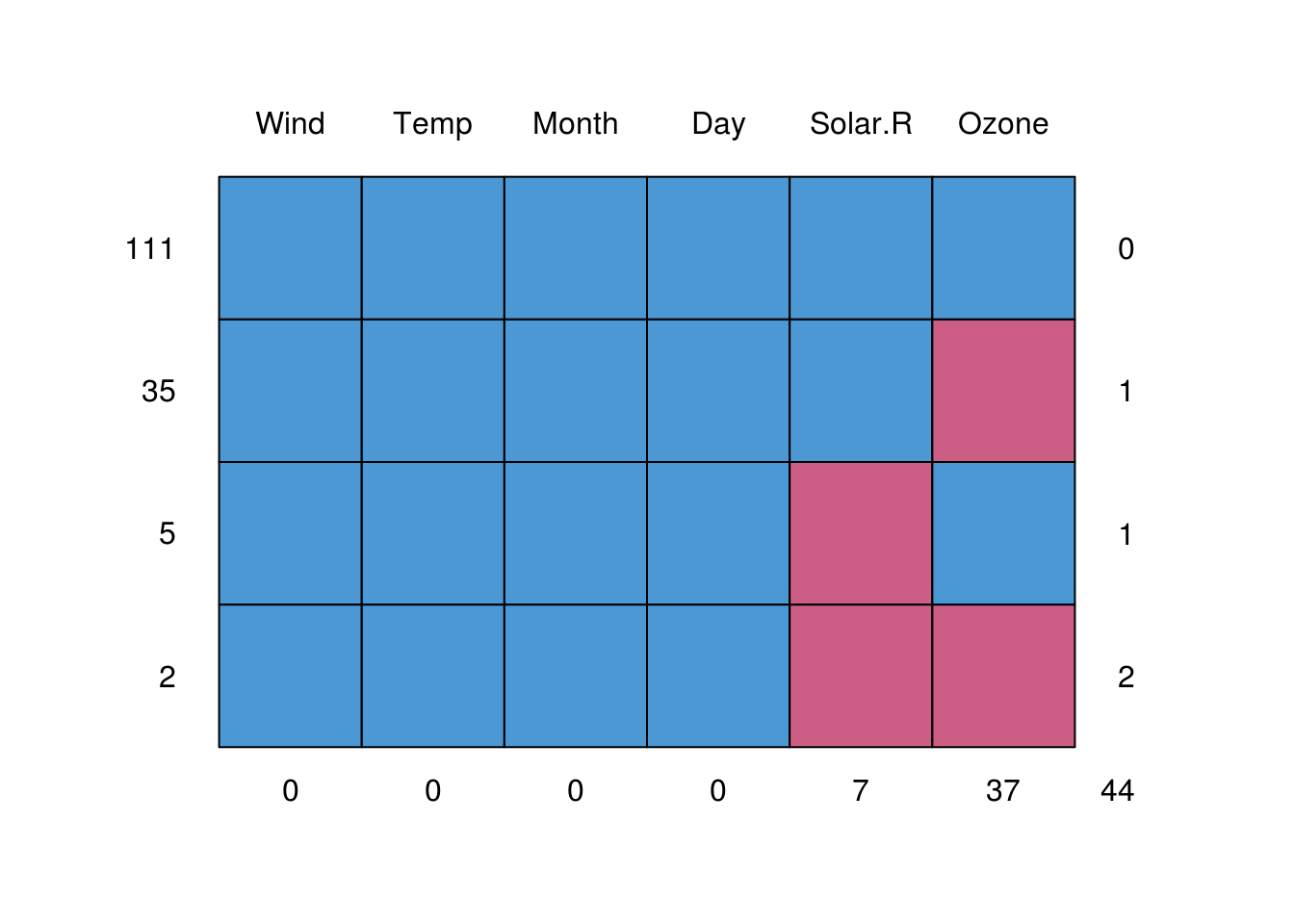
Wind Temp Month Day Solar.R Ozone
111 1 1 1 1 1 1 0
35 1 1 1 1 1 0 1
5 1 1 1 1 0 1 1
2 1 1 1 1 0 0 2
0 0 0 0 7 37 44In this table, md.pattern list the number of cases with particular patterns of missing data.
- Each row describes a misisng data ‘pattern’
- The first column indicates the number of cases
- The central columns indicate whether a particular variable is missing for the pattern (0=missing)
- The last column counts the number of values missing for the pattern
- The final row counts the number of missing values for each variable.
Visualising missingness
Graphics can also be useful to explore patterns in missingness.
rct.data contains data from an RCT of functional imagery training (FIT) for weight loss, which measured outcome (weight in kg) at baseline and two followups (kg1, kg2, kg3). The trial also measured global quality of life (gqol).
As is common, there were some missing data at the follouwp:
fit.data <- readRDS("data/fit-weight.RDS") %>%
select(kg1, kg2, kg3, age, gqol1)
mice::md.pattern(fit.data)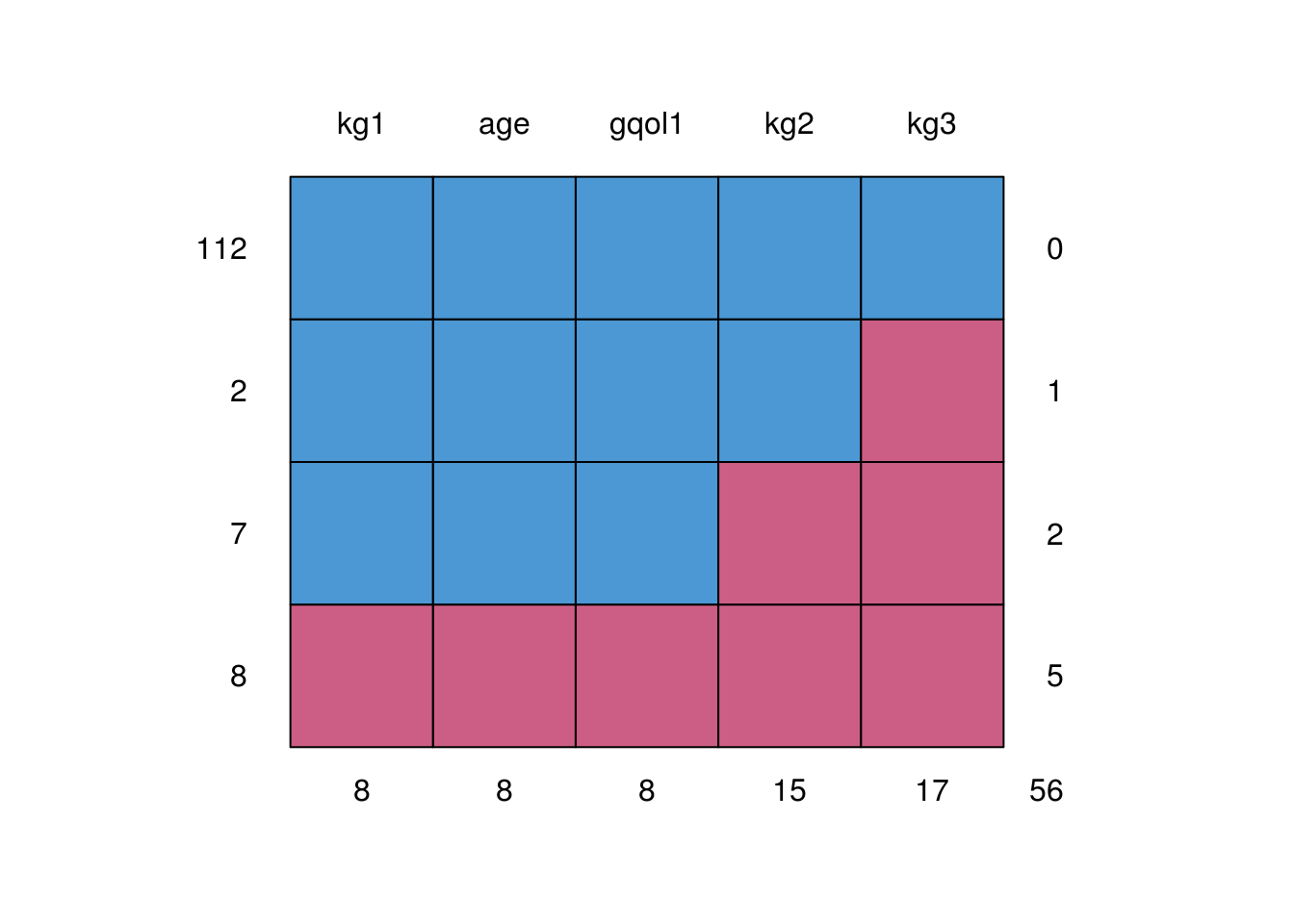
kg1 age gqol1 kg2 kg3
112 1 1 1 1 1 0
2 1 1 1 1 0 1
7 1 1 1 0 0 2
8 0 0 0 0 0 5
8 8 8 15 17 56We might be interested to explore patterns in which observations were missing. Here we use colour to identify missing observations as a function of the data recorded at baseline:
fit.data %>%
mutate(missing.followup = is.na(kg2)) %>%
ggplot(aes(kg1, age, color=missing.followup)) +
geom_point()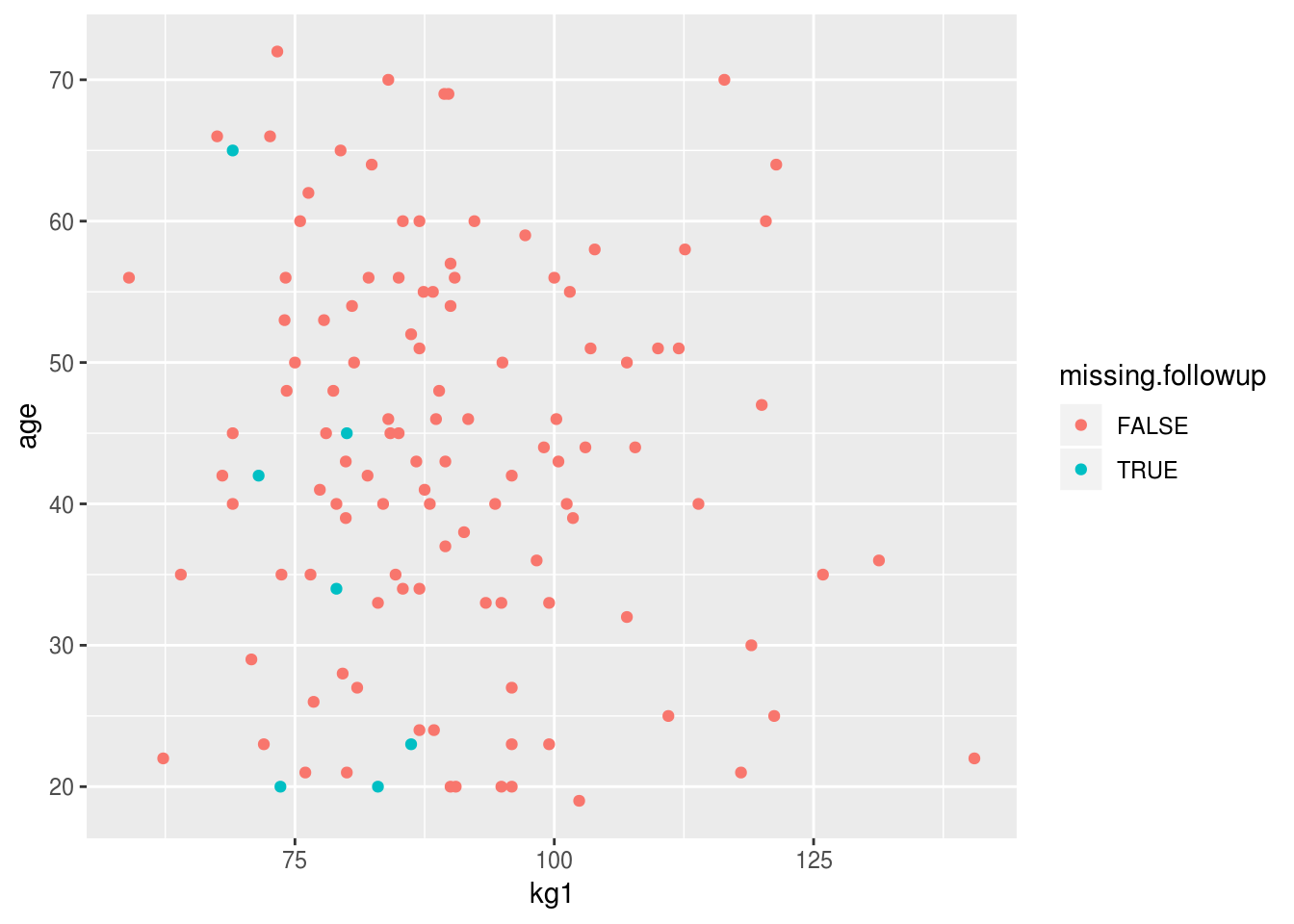
There’s a clear trend here for lighter patients (at baseline) to have more missing data at followup. There’s also a suggestion that younger patients are more likely to have been lost to followup.
If needed, we could perform inferential tests for these differences:
t.test(kg1 ~ is.na(kg2), data=fit.data)
Welch Two Sample t-test
data: kg1 by is.na(kg2)
t = 4.7153, df = 11.132, p-value = 0.000614
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
7.005116 19.236238
sample estimates:
mean in group FALSE mean in group TRUE
90.59211 77.47143
t.test(age ~ is.na(kg2), data=fit.data)
Welch Two Sample t-test
data: age by is.na(kg2)
t = 1.2418, df = 6.5246, p-value = 0.2571
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-7.39455 23.25169
sample estimates:
mean in group FALSE mean in group TRUE
43.50000 35.57143 However, given the small number of missing values and the post-hoc nature of these analyses these tests are rather underpowered and we might prefer to report and comment on the plot alone.
For some nice missing data visualisation techniques, including those for repeated measures data, see @zhang2015missing.