Checking assumptions
The text below continues on from this example of factorial Anova.
If we want to check that the assumptions of our Anova models are met, these tables and plots would be a reasonable place to start. First running Levene’s test:
car::leveneTest(eysenck.model) %>%
pander()| Df | F value | Pr(>F) | |
|---|---|---|---|
| group | 9 | 1.031 | 0.4217 |
| 90 | NA | NA |
Then a QQ-plot of the model residuals to assess normality:
car::qqPlot(eysenck.model)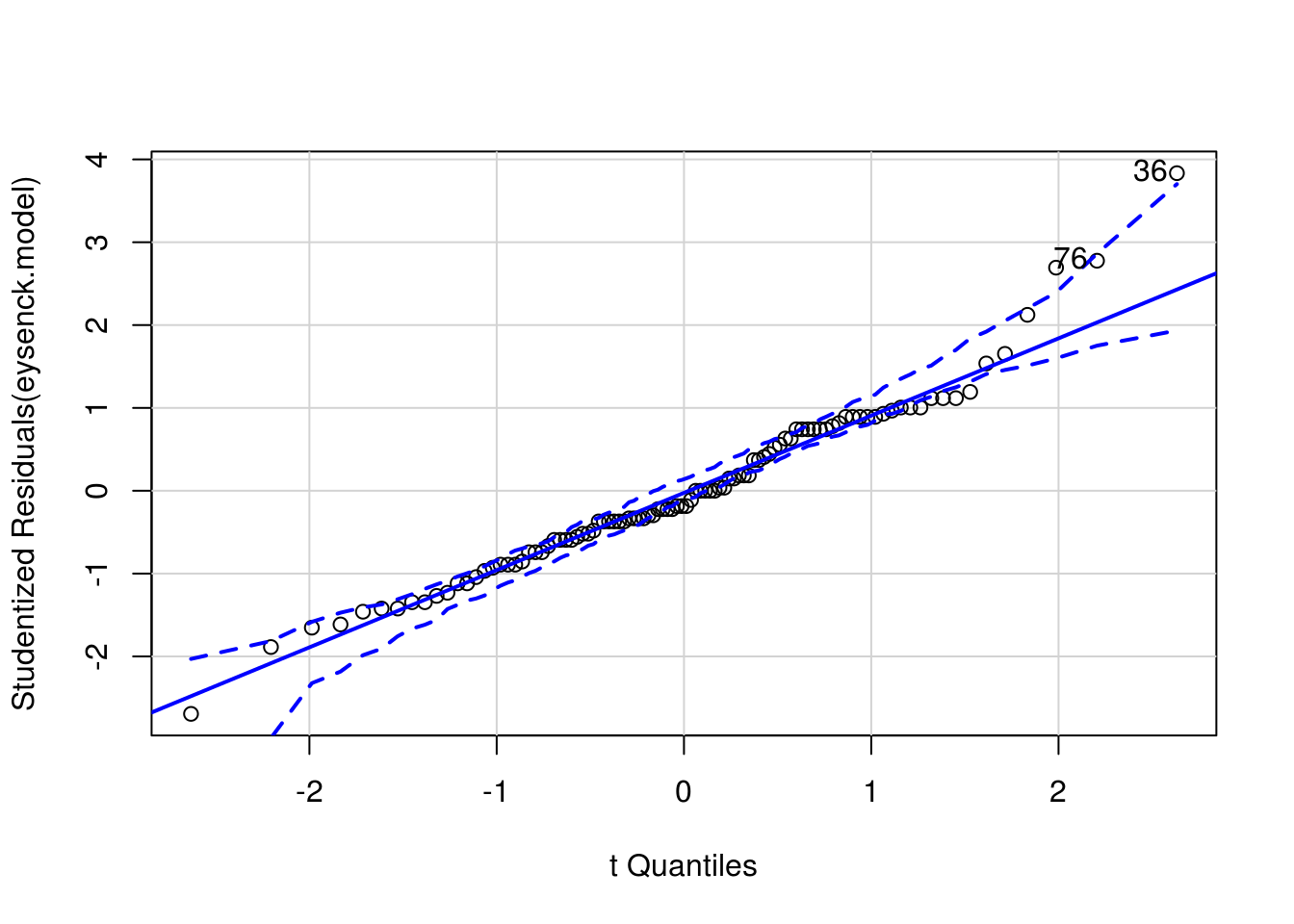
Figure 8.1: QQ plot to assess normality of model residuals
[1] 36 76And finally a residual-vs-fitted plot:
data_frame(
fitted = predict(eysenck.model),
residual = residuals(eysenck.model)) %>%
# and then plot points and a smoothed line
ggplot(aes(fitted, residual)) +
geom_point() +
geom_smooth(se=F)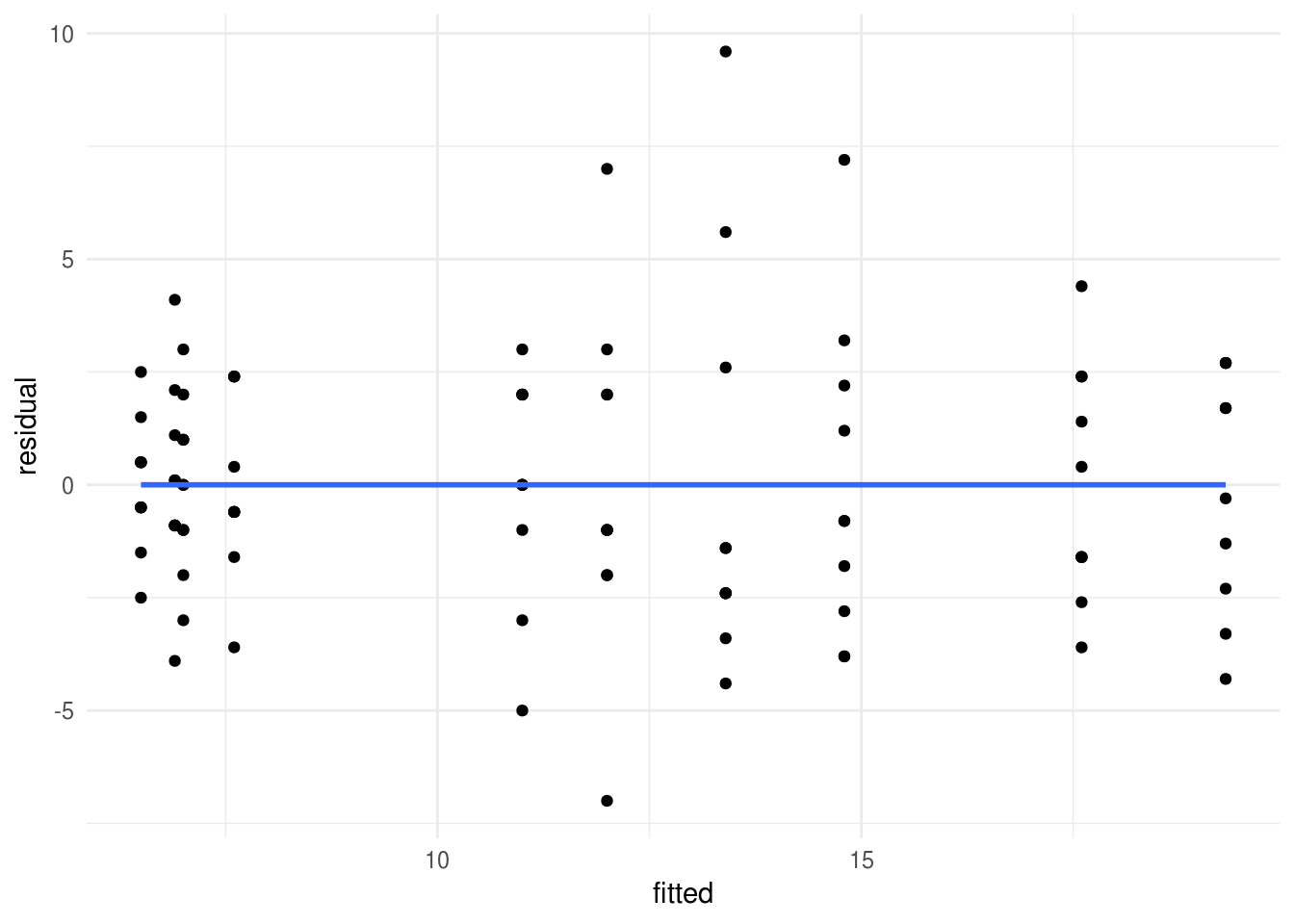
Figure 8.2: Residual vs fitted (spread vs. level) plot to check homogeneity of variance.
For more on assumptions checks after linear models or Anova see: http://www.statmethods.net/stats/anovaAssumptions.html